What Can Uni-Mol Do Too? | Demonstrating Excellent Predictive Performance in Tsinghua's MoleculeCLA Evaluation
AI-driven drug discovery relies on the accurate characterization of molecular features. The newly launched MoleculeCLA dataset by Professor Lanyan Yan's team at the Institute for Intelligent Industry, Tsinghua University, provides a new, multi-dimensional evaluation platform for molecular representation by generating large-scale computationally docked chemical, physical, and biological properties with no experimental noise. On this benchmark, Uni-Mol performed exceptionally well in end-to-end fine-tuning, achieving an average Pearson correlation coefficient of 0.68, ranking first among all pre-trained deep models and traditional molecular descriptors. This fully demonstrates its advantages in molecular feature extraction and prediction tasks. The preprint of the research paper entitled "MoleculeCLA: Rethinking Molecular Benchmark via Computational Ligand-Target Binding Analysis" has been published on arXiv.
MoleculeCLA Evaluation Process
The evaluation process of MoleculeCLA consists of three core stages to ensure comprehensive testing of the model in multi-dimensional property prediction tasks:
1.Docking Simulation
Perform standard chemical preprocessing and grid generation for approximately 140,000 structurally diverse small molecules and 10 representative protein targets (kinase ABL1, GPCR ADRB2, ion channel GluA2, nuclear receptor PPARG, epigenetic enzyme HDAC2, CYP2C9, SARS-CoV-2 3CL protease, HIV protein, KRAS, PDE5);
Use Schrödinger Glide for high-throughput molecular docking to obtain stable and reproducible binding conformations.
2.Property Extraction
Extract 9 key indicators from the docking outputs:
Chemical properties: hydrophobicity (lipo), hydrogen bond formation tendency (hbond);
Physical properties: van der Waals energy (evdw), Coulomb energy (ecoul), polar site contribution (esite), rotatable bond energy (erotb), internal torsion energy (einternal);
Biological properties: docking score (docking_score), model energy (emodel);
These properties cover chemical, physical, and biological aspects, enabling in-depth assessment of the model's ability to characterize molecular-target interactions.
3.Model Fine-Tuning Methods
Linear Probe: Freeze the pre-trained encoder and only train the linear regression head to quickly measure the linear separability of features;
MLP fine-tuning: Attach a small multi-layer perceptron after the encoder output to test the gain effect of simple non-linear models;
Fine-Tune (full-parameter fine-tuning): Perform end-to-end training on all parameters of the model to test the ultimate performance in real downstream tasks.
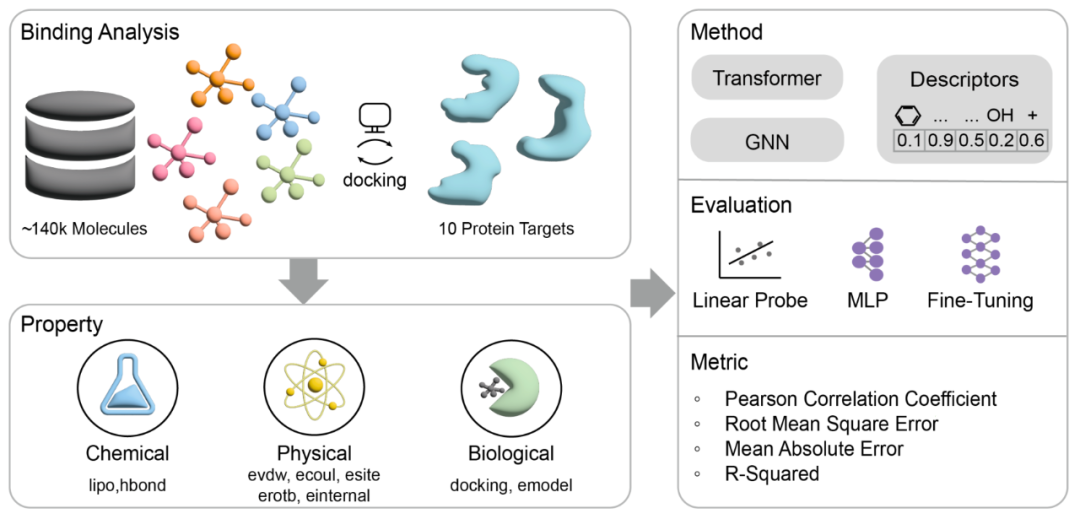
Figure 1 Overview of the MoleculeCLA method. The full process from molecular-target docking, to the extraction of three types of properties, to the three model fine-tuning methods: Linear Probe, MLP, and Fine-Tune.
Introduction to MoleculeCLA Dataset Benchmark
The following is the specific composition and splitting strategy of the MoleculeCLA dataset:
1.Selection of Molecules and Targets
140,000 structurally diverse small molecules: sourced from commercial compound libraries, ensuring diversity through chemical fingerprinting and clustering;
10 representative protein targets: covering kinases (ABL1), GPCRs (ADRB2), ion channels (GluA2), nuclear receptors (PPARG), epigenetics (HDAC2), CYP2C9, SARS-CoV-2 3CL protease, HIV protein, KRAS, PDE5, catering to both classical and emerging drug discovery needs.
2.Data Splitting Strategy
Adopting scaffold split: Divide the dataset into training, validation, and test sets based on molecular scaffolds to ensure the chemical structural independence of the three and avoid data leakage;
Scale: 112,557 training samples, 14,070 validation samples, and 14,070 test samples, ensuring large-scale model training while fully evaluating generalization ability.
3.Chemical Space and Property Diversity
Chemical coverage: Comparable to the PCBA dataset in t-SNE visualization of chemical space, but with a scale of only one-third, reflecting efficient representativeness;
Property dimensions: The 9 extracted properties generally have low correlation (|r| < 0.3), forcing the model to perform well in the three dimensions of chemistry, physics, and biology.
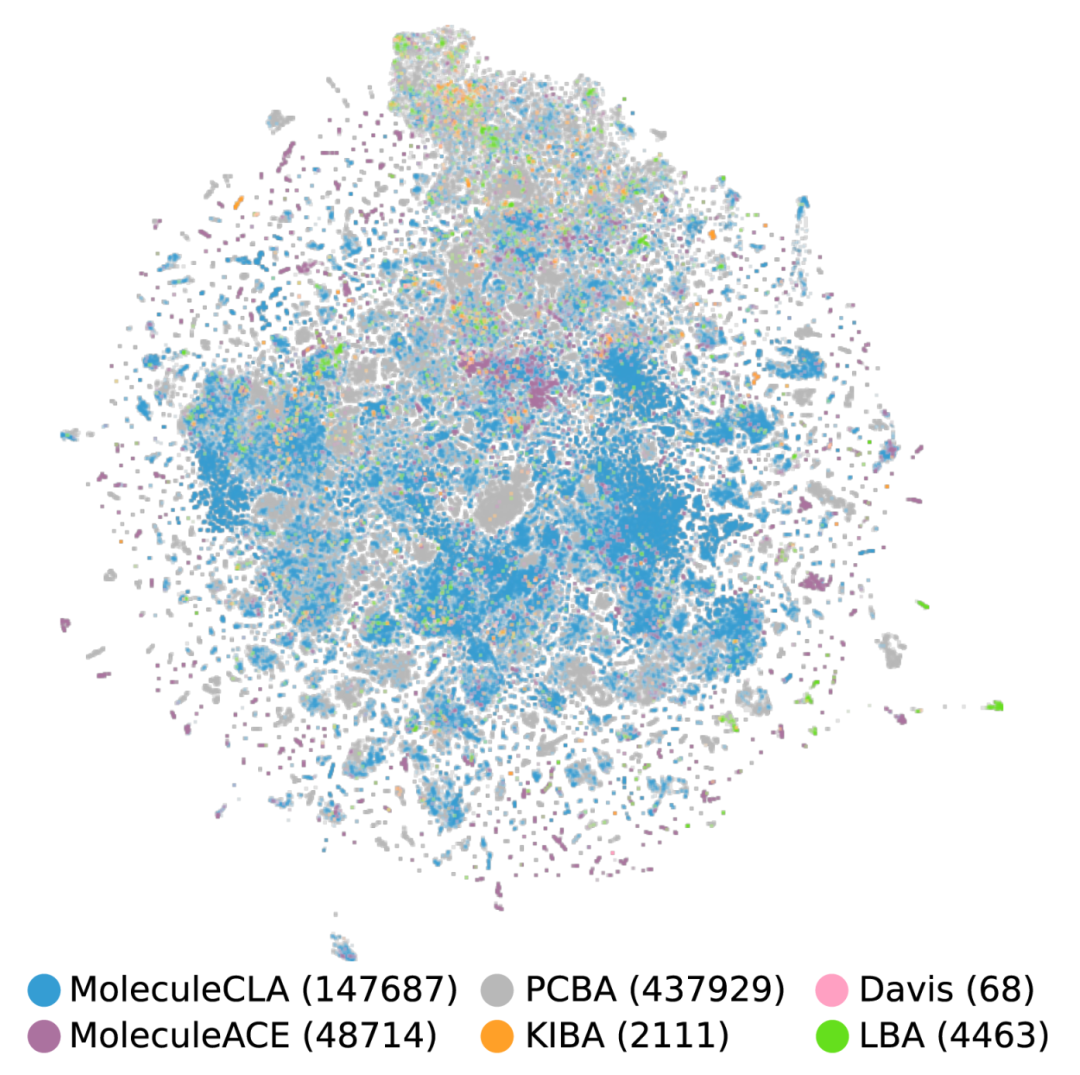
Figure 2 t-SNE visualization of chemical space covered by MoleculeCLA (147,687) and other datasets (PCBA (437,929), Davis (68), MoleculeACE (48,714), KIBA (2,111), LBA (4,463)).
Model Fine-Tuning Methods and Uni-Mol's Performance
On MoleculeCLA, we adopted three model fine-tuning methods: Linear Probe, MLP fine-tuning, and full-parameter fine-tuning (Fine-Tune) to systematically evaluate the model's performance in 9 chemical/physical/biological property prediction tasks.
1.Linear Probe
Linear Probe test involves freezing the pre-trained encoder and only training the linear regression head to evaluate the linear separability of latent vectors. If the linear head can achieve good results, it indicates that the pre-trained features already contain key information required for downstream tasks, without relying on complex non-linear heads or full-parameter fine-tuning. Among them, Uni-Mol achieved an average Pearson correlation coefficient of 0.582, proving that its pre-training stage has efficiently captured the linear laws of molecular-target interactions. However, traditional molecular descriptors (Descriptor2D/2D&3D) had average Pearson coefficients of 0.646 and 0.647, respectively, still leading all deep models, indicating that manually designed molecular fingerprints still have irreplaceable advantages under the "zero fine-tuning" condition.
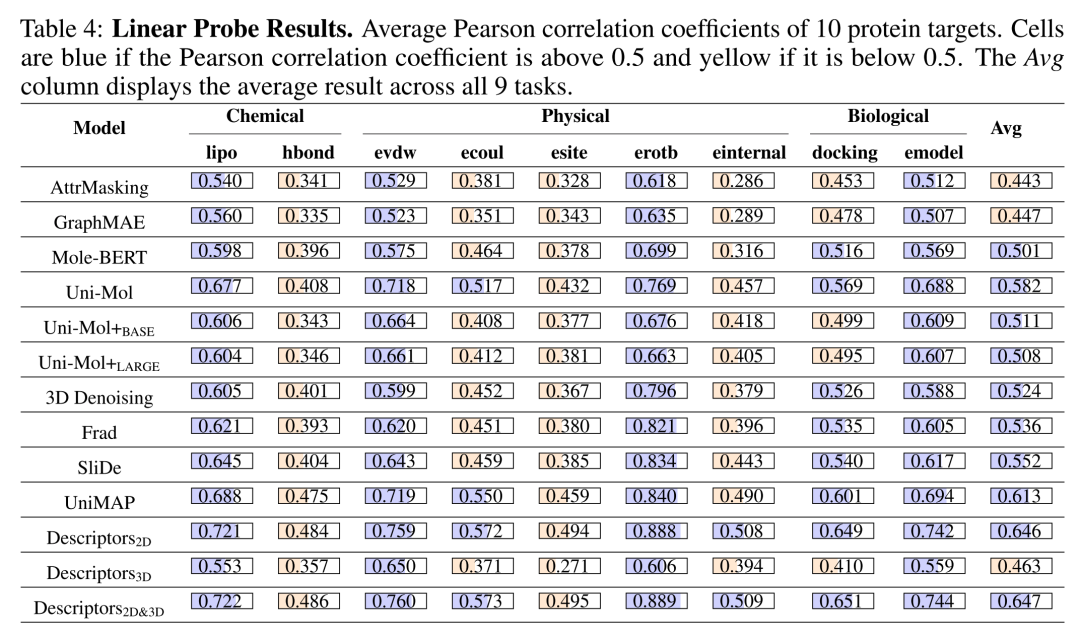
Table 1 Linear Probe Results. Average Pearson correlation coefficients of 10 protein targets. Cells are blue if the Pearson correlation coefficient is above 0.5 and yellow if it is below 0.5. The Avg column displays the average result across all 9 tasks.
2.Fine-Tune (Full-Parameter Fine-Tuning)
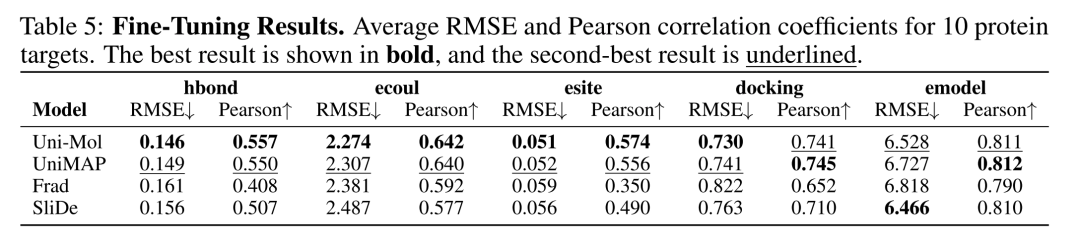
Fine-Tune refers to end-to-end training of all model parameters to meet the ultimate performance requirements of real tasks. Uni-Mol achieved an average Pearson correlation coefficient of 0.68 for 5 downstream tasks, ranking first in the tasks of hydrogen bond formation tendency (hbond), electrostatic interaction (ecoul), and polar site contribution (esite), and second in the prediction tasks of molecular docking affinity (docking) and model energy (emodel).
Table 2 Fine-Tuning Results. Average RMSE and Pearson correlation coefficients for 10 protein targets. The best result is shown in bold, and the second-best result is underlined.
3.Drug-Target Interaction (DTI)
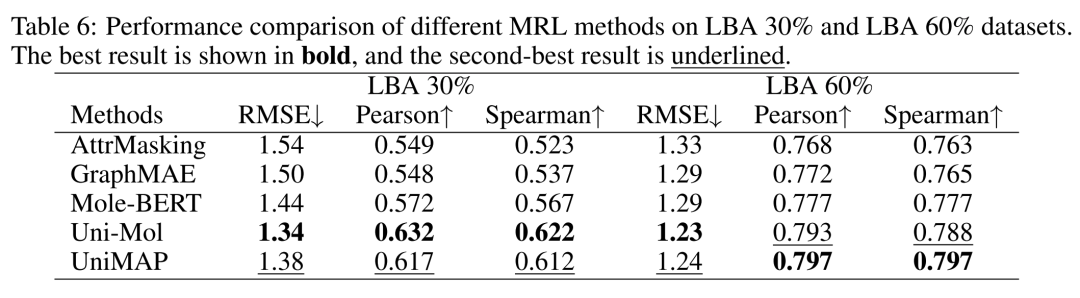
The drug-target interaction experiment embeds the pre-trained model into BindNet to realize end-to-end fine-tuning for PDBBind ligand binding affinity (LBA) prediction tasks, using a 30%/60% data split. In the LBA 30% task, Uni-Mol's RMSE, Pearson coefficient, and Spearman coefficient were comprehensively better than other models; in the LBA 60% task, Uni-Mol's RMSE of 1.23 was better than other models, and its Pearson and Spearman coefficients were second only to the best model. Although the MoleculeCLA benchmark is generated by computational simulation, it can effectively predict binding affinity in real experiments; Uni-Mol's excellent performance in the DTI task verifies the wide applicability and reliability of its pre-training and fine-tuning strategies.
Table 3 Drug-Target Interaction Results. The LBA task dataset split uses 30%/60%.
Through these three rounds of tests, Uni-Mol has shown leading performance in fine-tuning and drug-target interaction tasks, verifying its advantages in molecular representation and property prediction. As a pre-trained model, Uni-Mol can exhibit excellent predictive performance after fine-tuning due to the following points:
3D prior alignment: Uni-Mol's pre-training tasks enhance sensitivity to three-dimensional geometry and force field features, naturally fitting force field-related tasks in MoleculeCLA such as Coulomb energy (ecoul) and van der Waals energy (evdw);
Large-scale coverage: Massive conformation samples and diverse chemical spaces in the pre-training stage enable the model to quickly adapt to the multi-dimensional property distribution of 140,000 molecules during fine-tuning;
High-capacity architecture: The Transformer architecture has both long-range dependency and local chemical bond information capture capabilities, and multi-scale feature fusion helps to finely fit downstream tasks;
Two-stage paradigm: First, learn general representations in massive data in a "breadth-first" manner, then "deeply focus" on specific tasks, and amplify pre-training advantages through Fine-Tune to achieve the best balance of "pre-training + fine-tuning".
Summary
On MoleculeCLA, a new large-scale, experimental noise-free benchmark, Uni-Mol has achieved stable and leading results in end-to-end fine-tuning, demonstrating its generalization ability and practical value in molecular property prediction tasks.